Tutorial to design gRNA for CRISPR base editing knockout system.¶
Step 1: Prepare input file¶
CRISPR-BETS allows Genbank, Snapgene, Fasta file formats as input.
- Genbank
The GenBank format for most genes can be downloaded from NCBI(https://www.ncbi.nlm.nih.gov/).
Download
Genbank example file.
- Snapgene
For Snapgene file(.dna), CRISRP-BETS uses a continuous CDS feature to determine the coding sequence of the gene, and different continuous CDS features serve as different isoforms. Snapgene file(.dna) could be generated by SnapGene software (from Insightful Science; available at https://www.snapgene.com/).
Download
Snapgene example file.
- Fasta
The CRISPR-BETS will use exonerate(https://www.ebi.ac.uk/about/vertebrate-genomics/software/exonerate) align CDS sequence to DNA sequence to found Splicing site.
Download
DNA Fasta example file. DownloadCDS Fasta example file
Step 2: Select and upload file¶
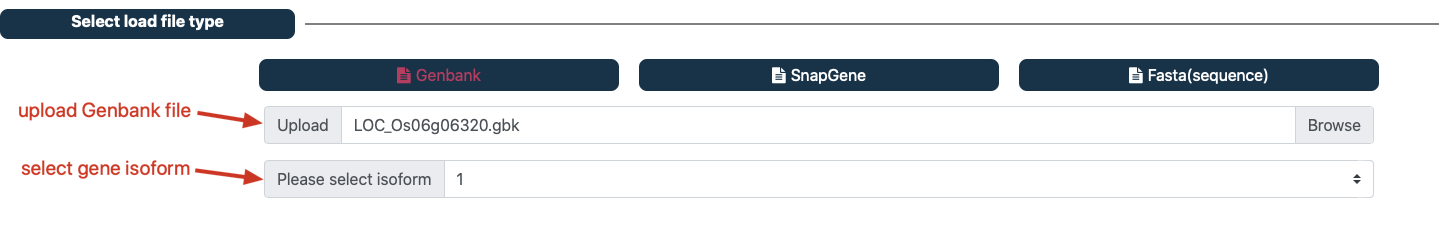
① upload GenBank or Snapgene file¶
Navigate to the ‘Select load file type’ panel. click ‘GenBank’ button (GenBank is the default input file).
Upload the GenBank files.
Select gene isoform (support for Genbank and Snapgene file).
② upload fasta file or sequences¶
Navigate to the ‘Select load file type’ panel. click ‘Fasta(sequence)’ button
Fill the text box with both the DNA and CDS sequences of a gene.
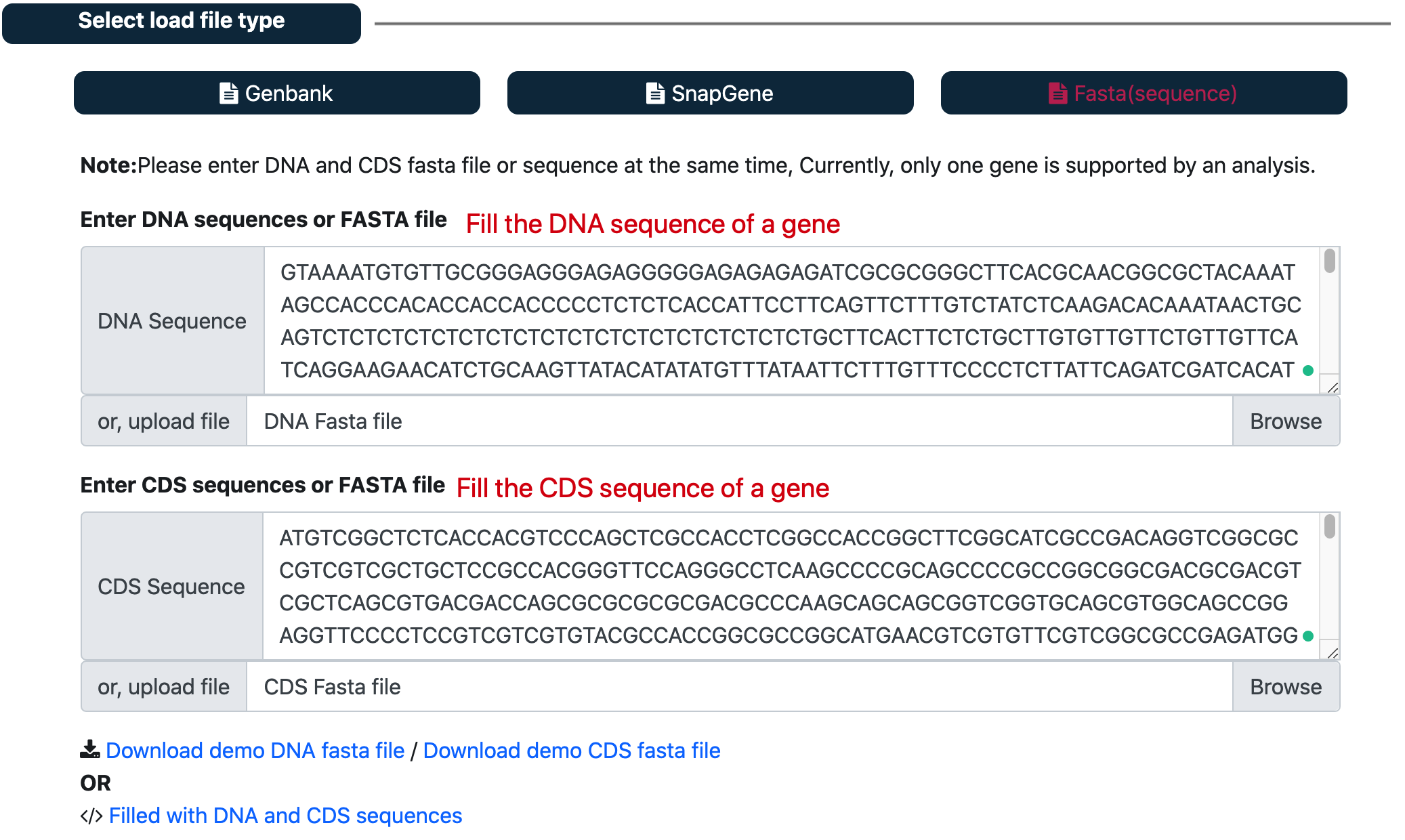
NOTE:In order to design cross-CDS gRNA, enter both DNA and CDS FASTA files or sequences. Currently, only one gene is supported by an analysis.
Step 3: Select edit system option¶
Navigate to the ‘Edit system option’ panel.
(optional) Select ‘Cas9 variants’, ‘PAM’ and ‘gRNA length’ will be automatically filled.
(optional) Select ‘Deaminase’, ‘Edit window’ will be automatically filled.
The user fill ‘PAM’, ‘gRNA length’ and ‘Edit window’.
(optional) The user select genome to estimate the specificity of each gRNA.
Click the ‘Analysis’ button and wait for the analysis to complete.
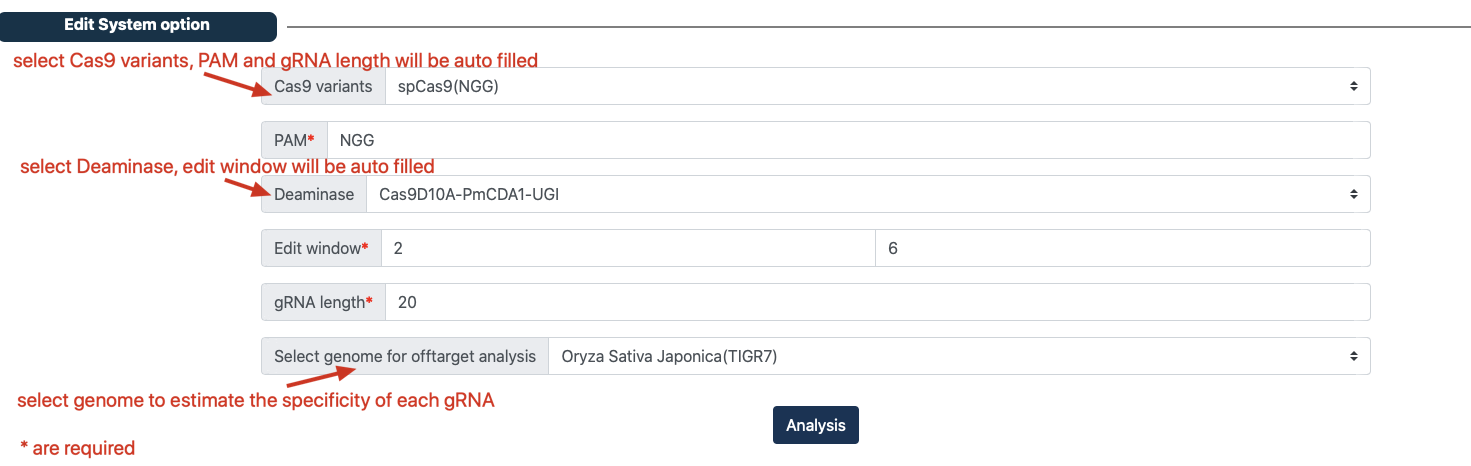
Parameter description¶
1. Cas9 variants: Select the appropriate Cas9 variant. Once selected, the parameters PAM and gRNA length will be filled in automatically, not required.
2. PAM: The default value is NGG, which can be customized, required.
3. Edit window: The default values are 1 and 20 (The farthest first base of PAM is represented as a 1), which can be customized, required.
4. gRNA length: The default value is 20, which can be customized, required.
5. Select genome for offtarget analysis: Select species genome for off-target analysis, this step will increase the running time of CRISPR-BETS, not required.
Step 4: Scanning result panel and download result information¶
Navigate to the ‘result panel’, the user can drag and zoom to check the detailed information of each gRNA.
Click ‘save result as txt’ button to download result.
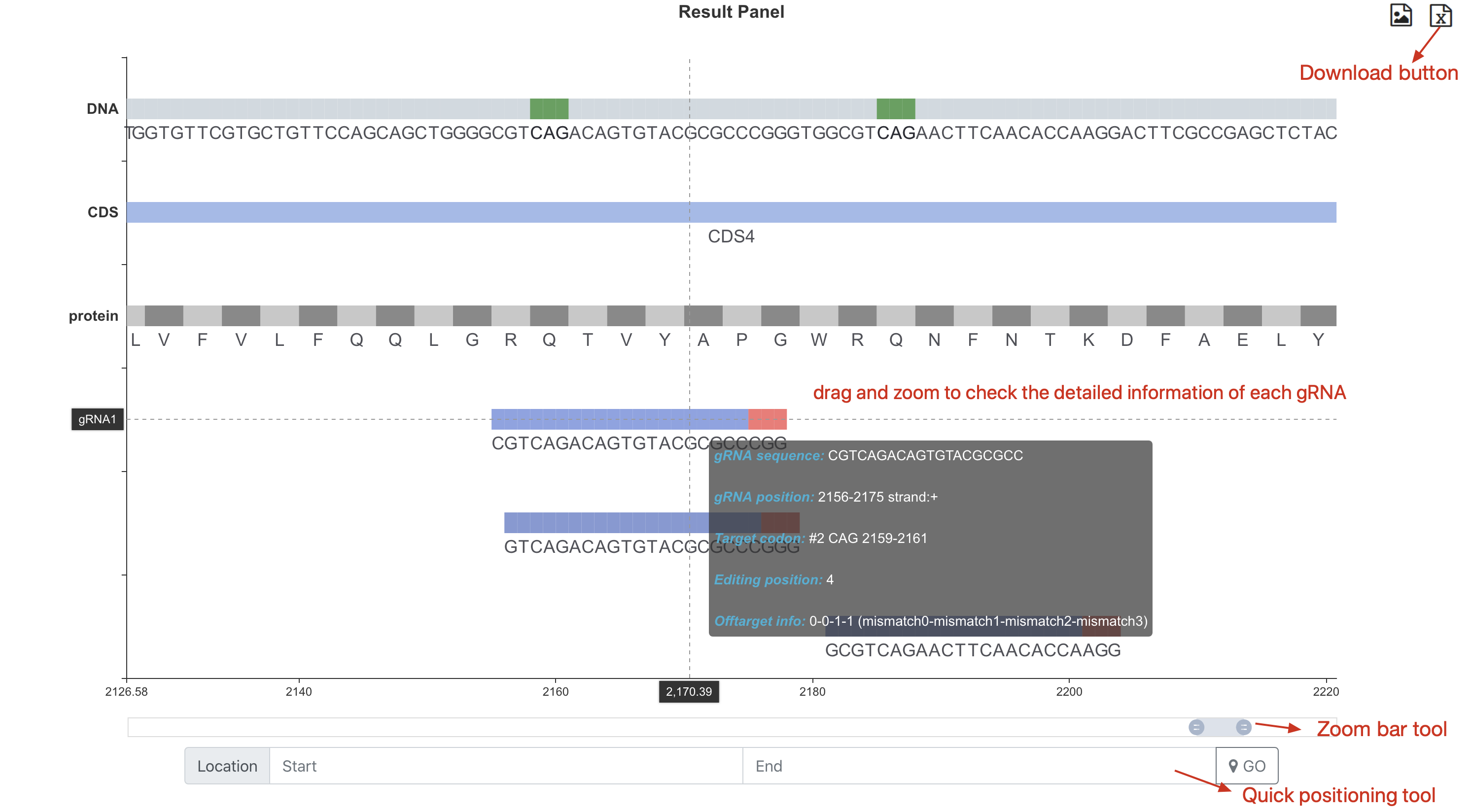
The following two functional buttons exist in the upper right corner of the results panel¶
Screenshot: Take a screenshot of the current page.

save result as file: Take a screenshot of the current page.

Output result description¶
The first row: Target gene name, determined according to the uploaded file name.

The second row: Edit system option.

The third row: Gene DNA sequences.

The fourth row: The position of CDS in DNA.

Row five to the last: Each target codon and its corresponding gRNA, different information is separated by ‘;’.
